Article Outline
検査陽性者数(チャーター便を除く国内事例)と政府の対応(新型コロナウイルス:Coronavirus)
(使用するデータ)
東洋経済オンライン:https://raw.githubusercontent.com/kaz-ogiwara/covid19/master/data/data.json
北海道・東北
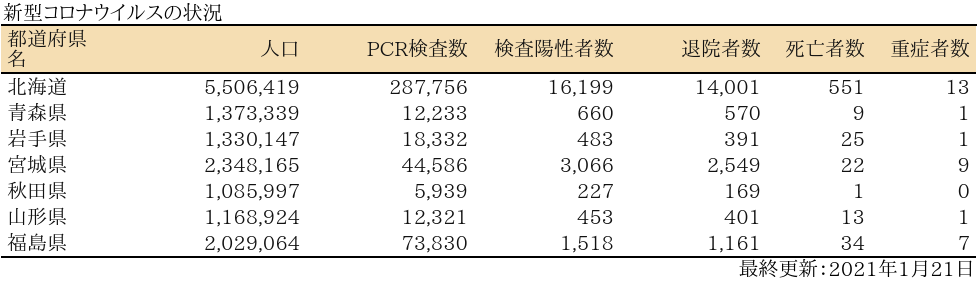
表
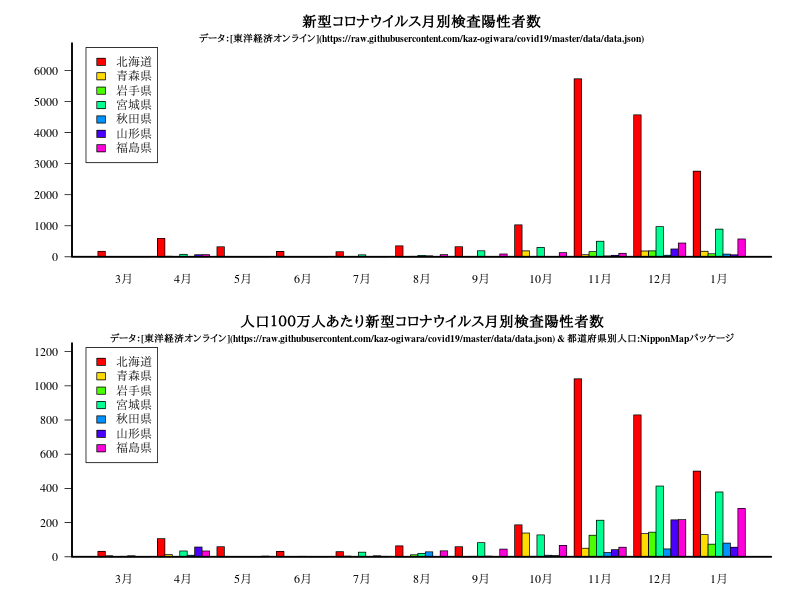
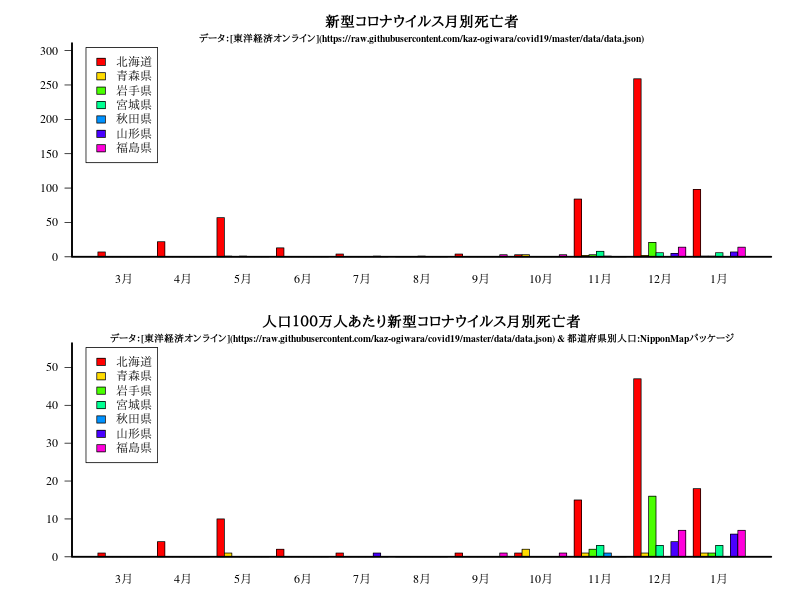
グラフ
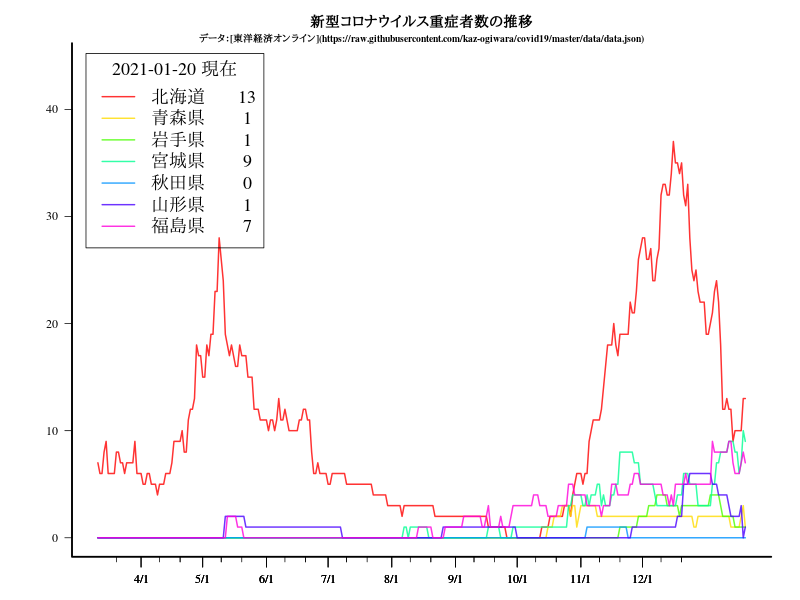
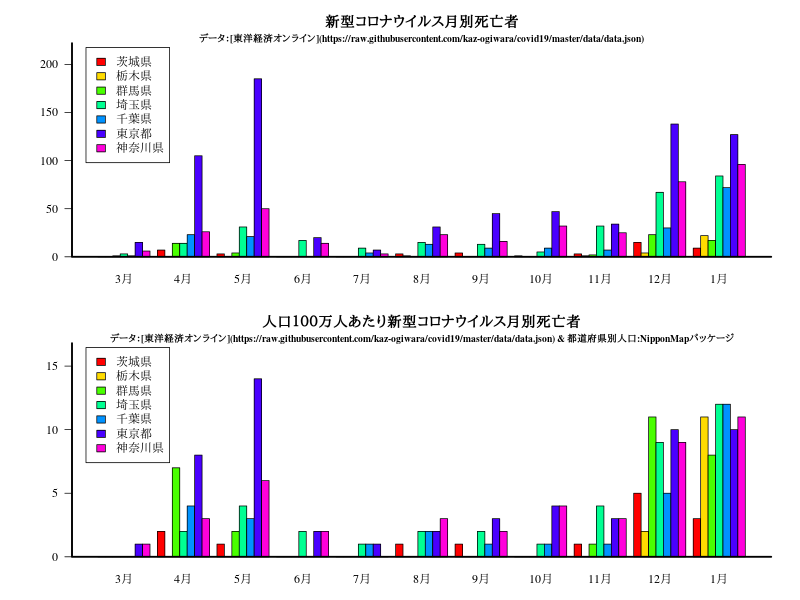
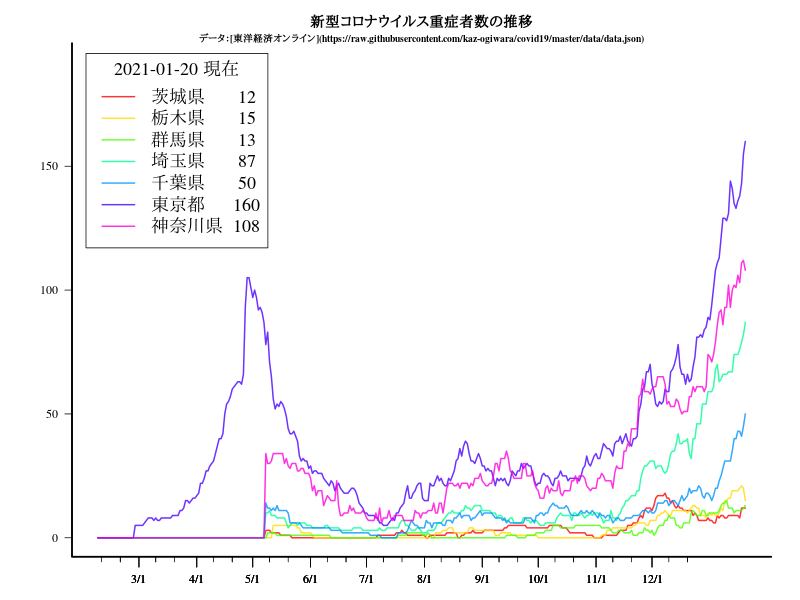
関東
表
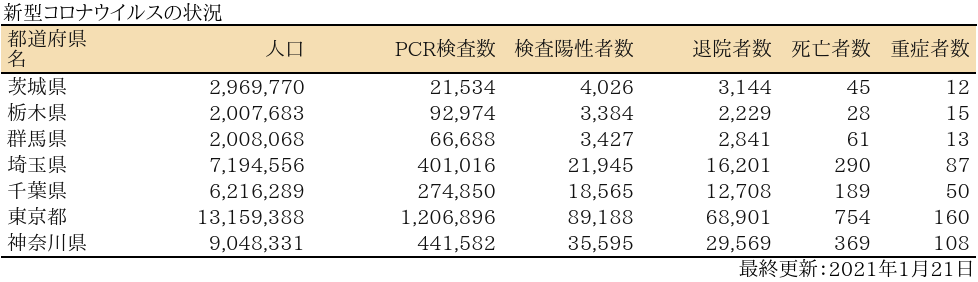
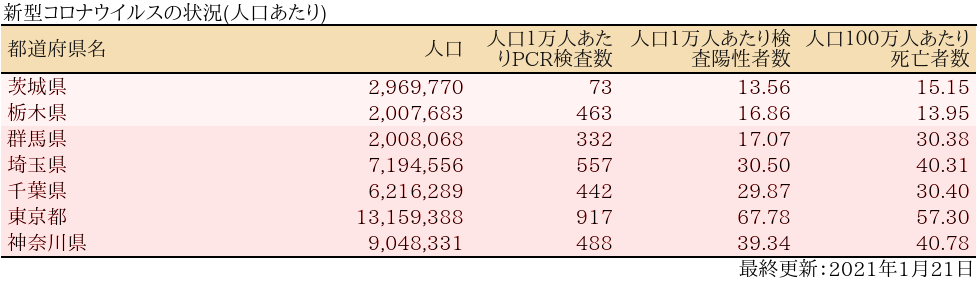
グラフ
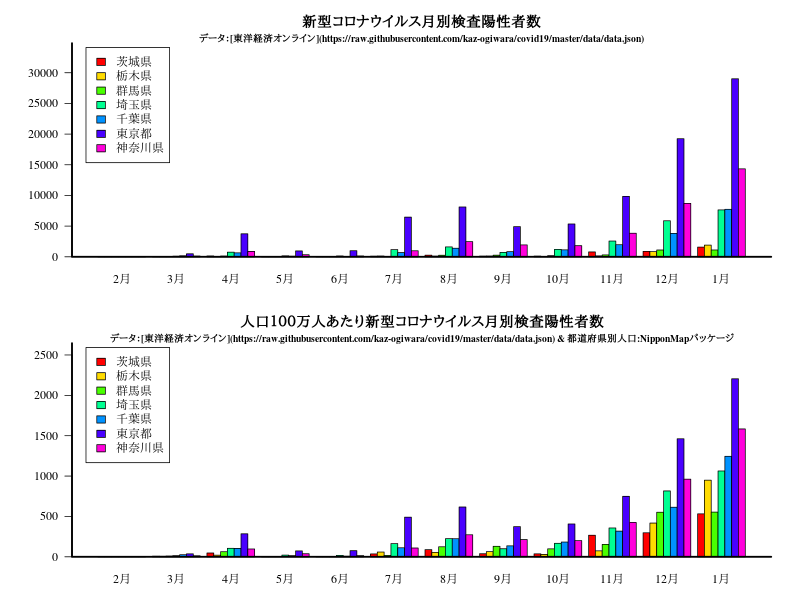
中部
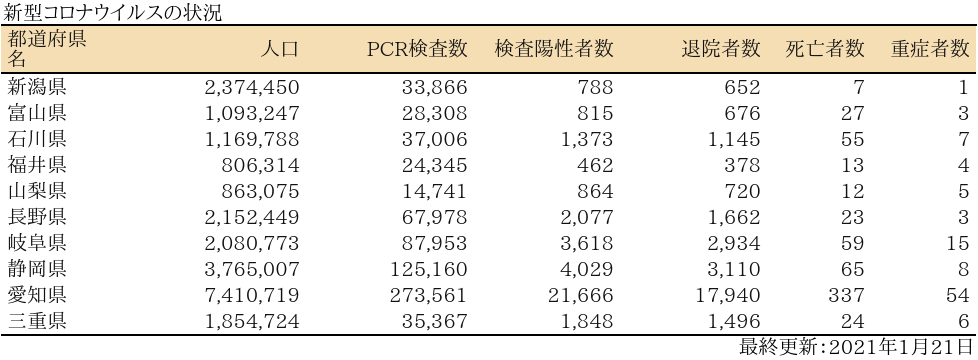
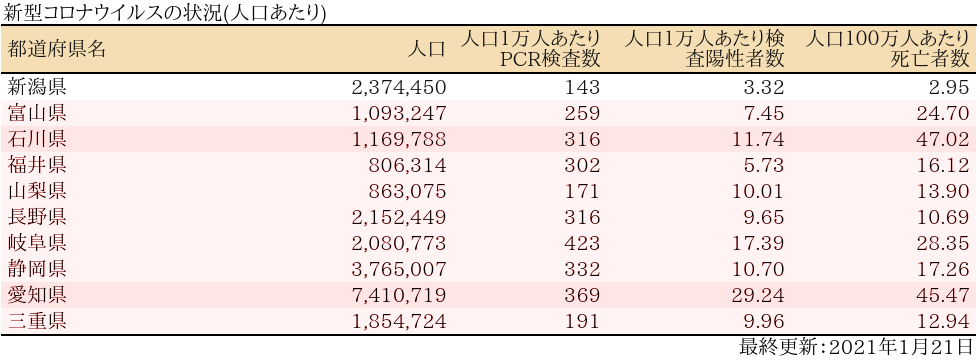
表
グラフ
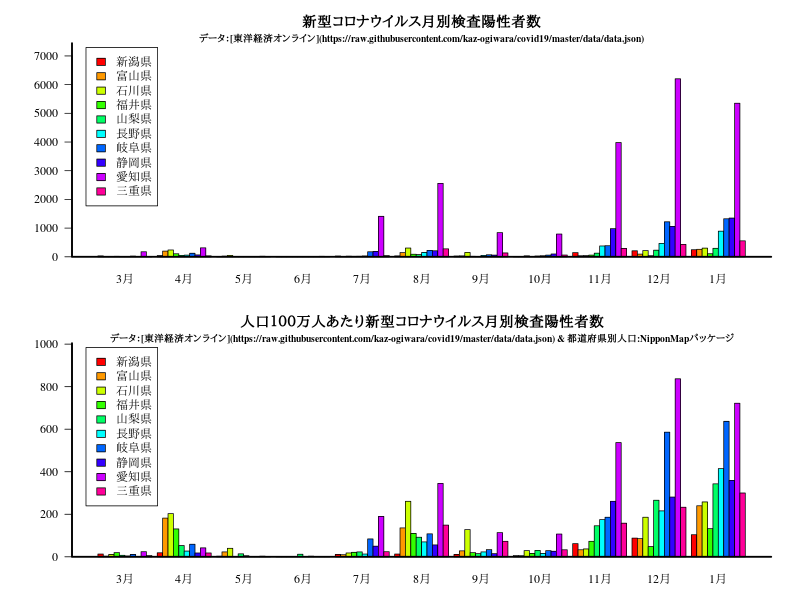
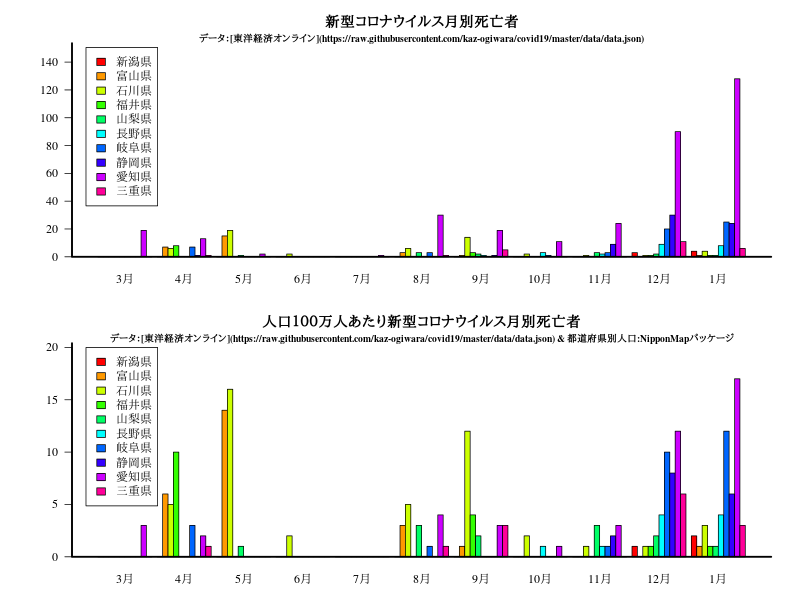
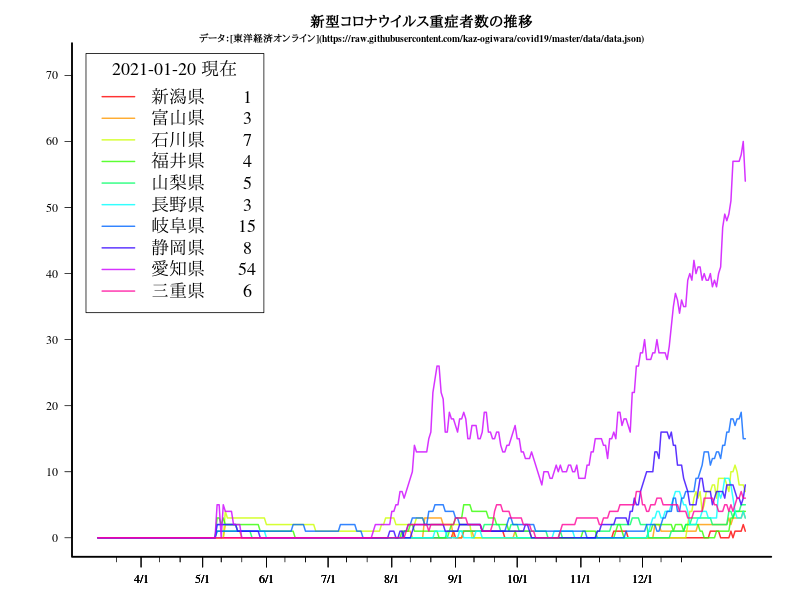
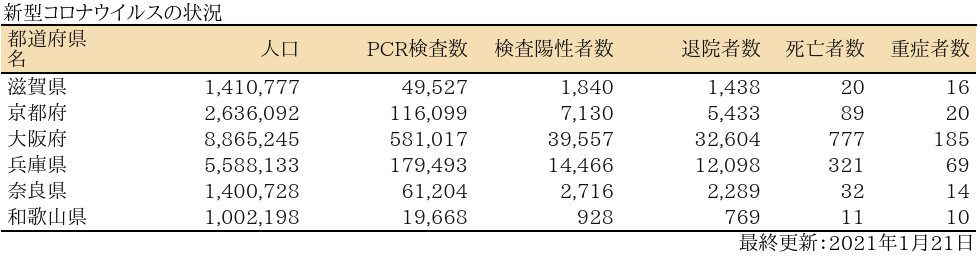
近畿
表
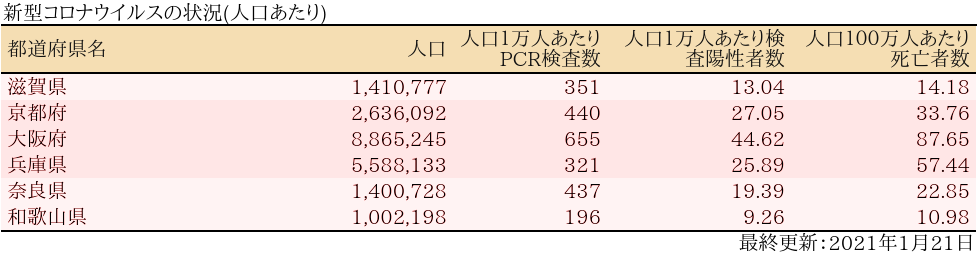
グラフ
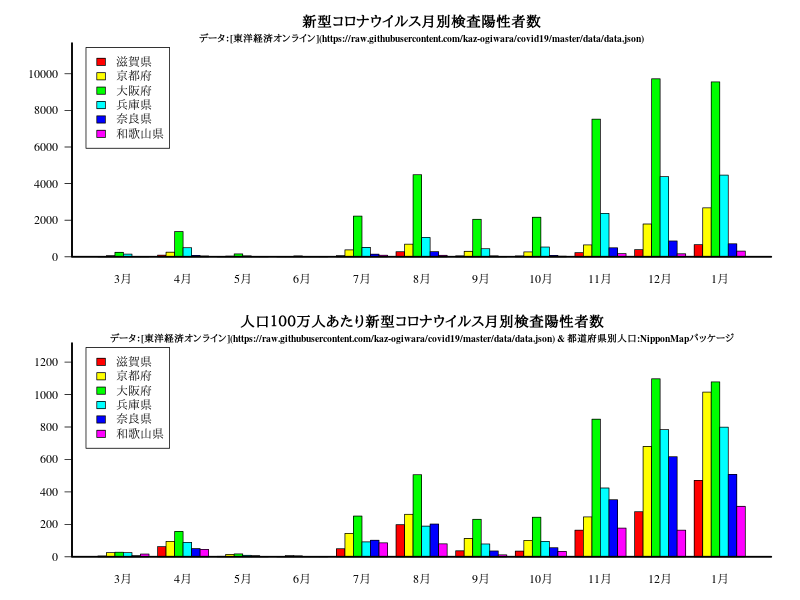
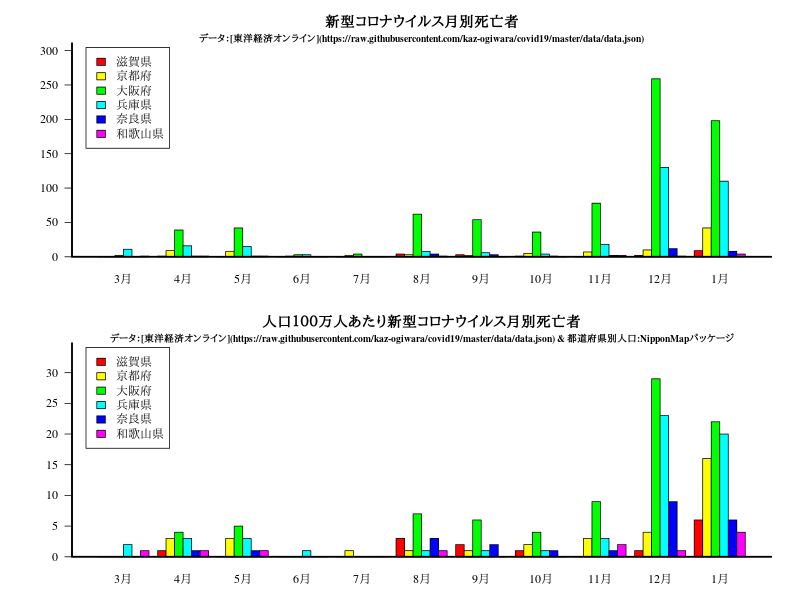
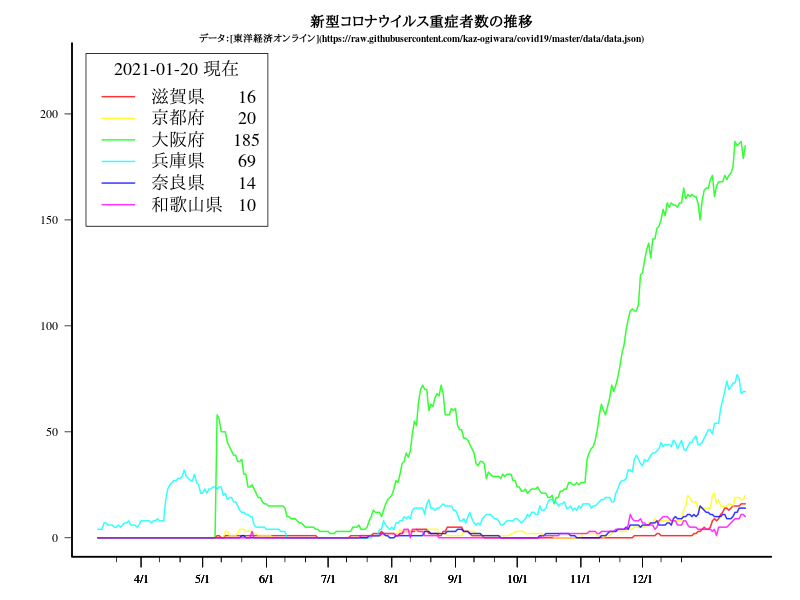
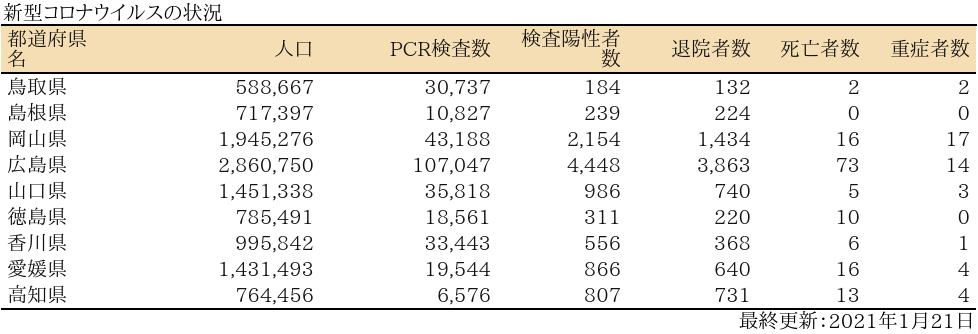
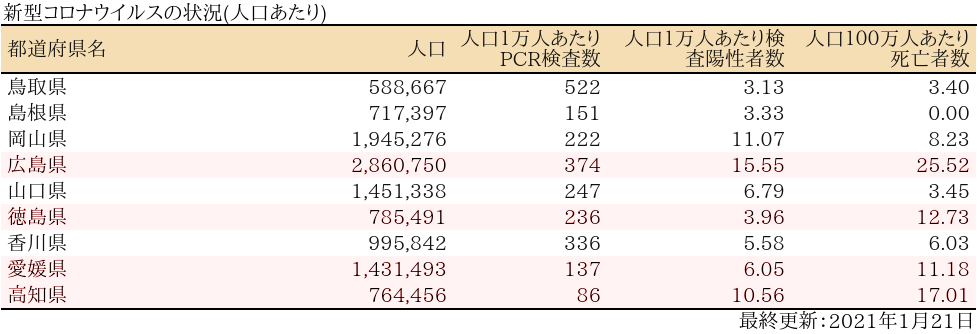
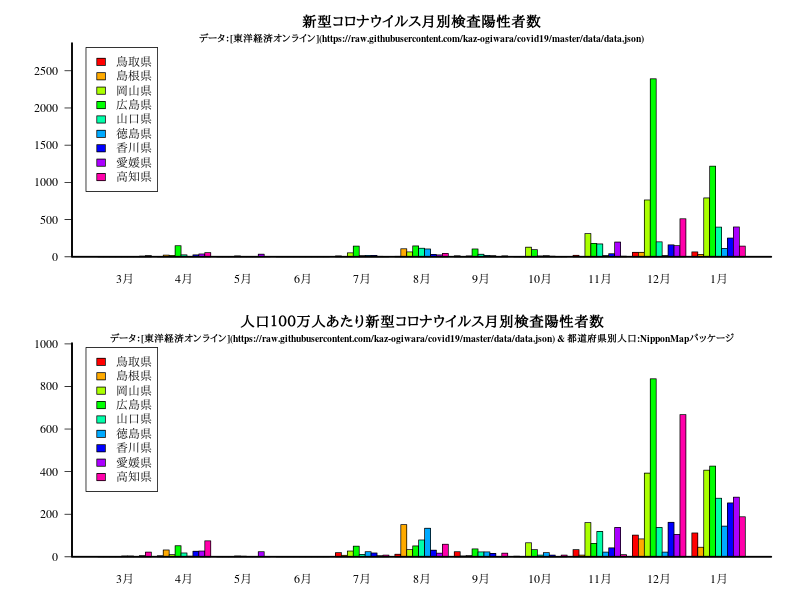
中国・四国
表
グラフ
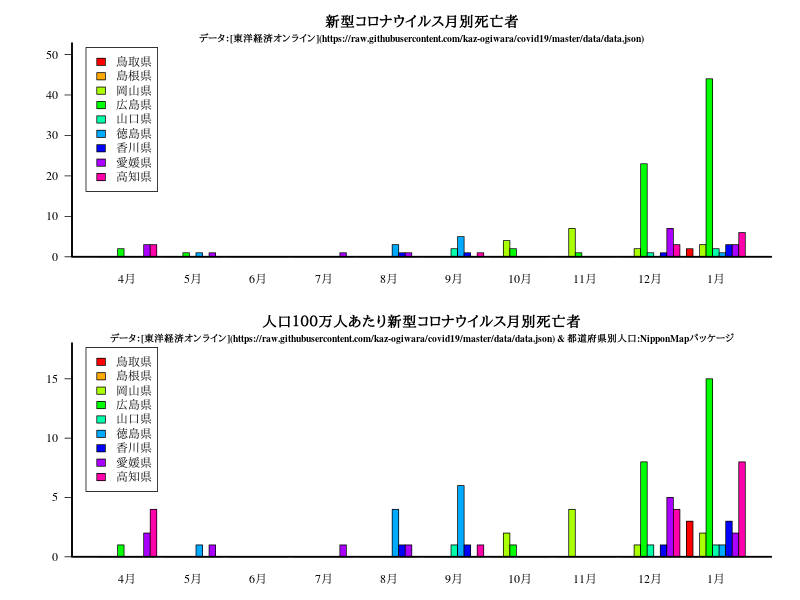
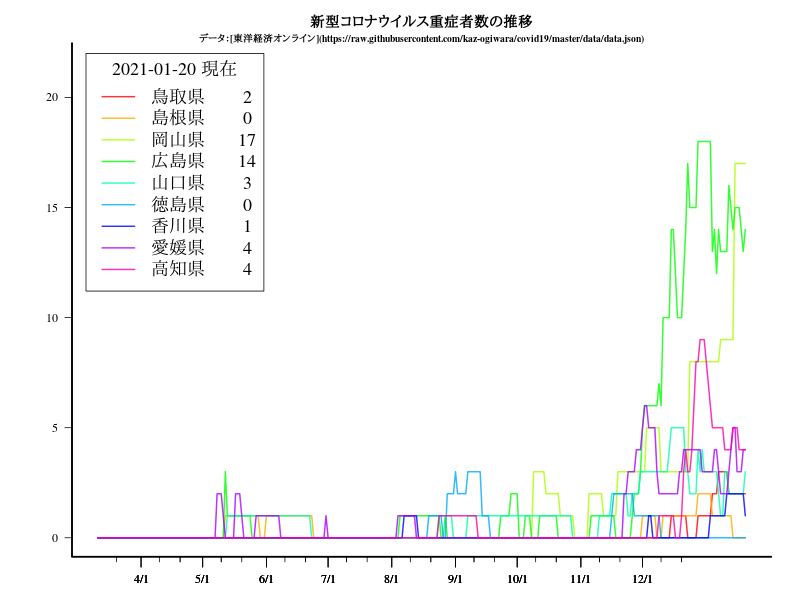
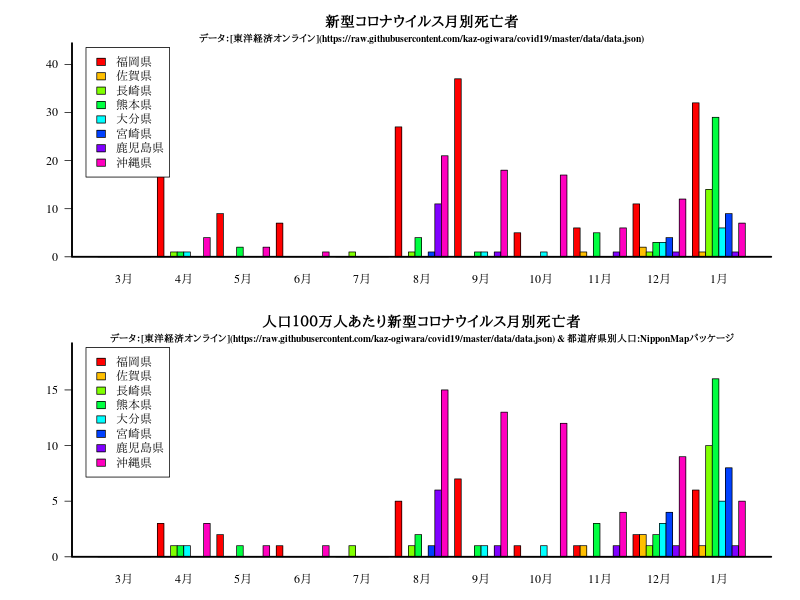
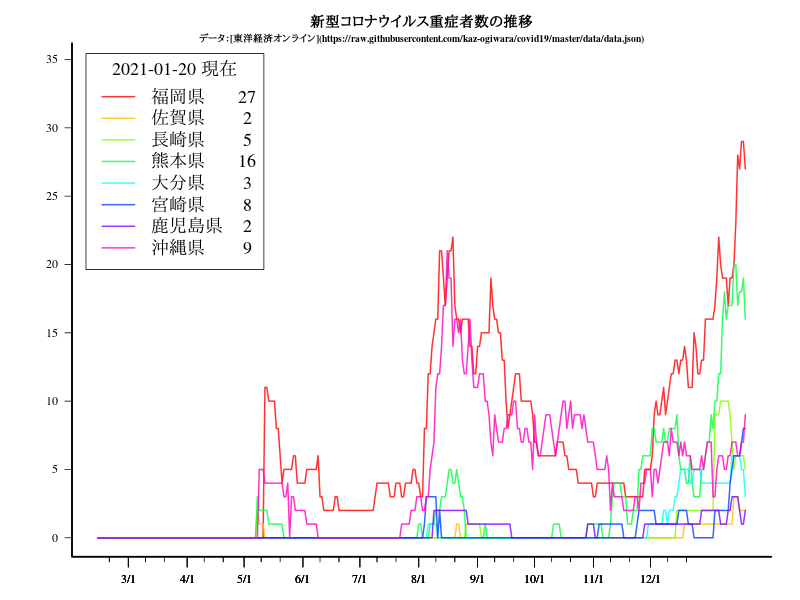
九州・沖縄
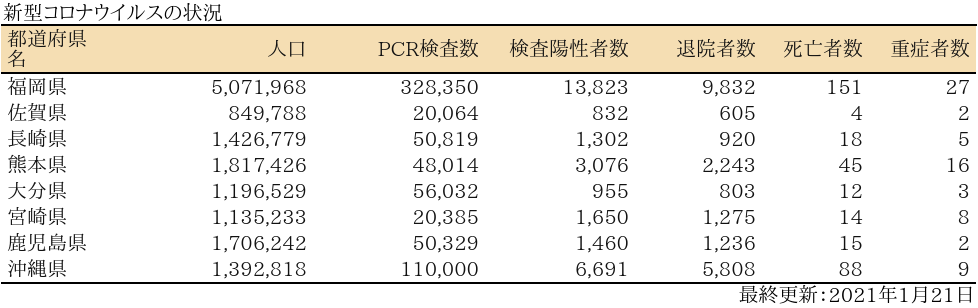
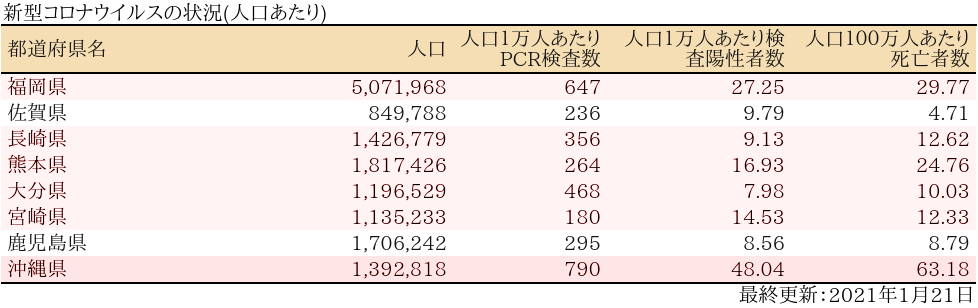
表
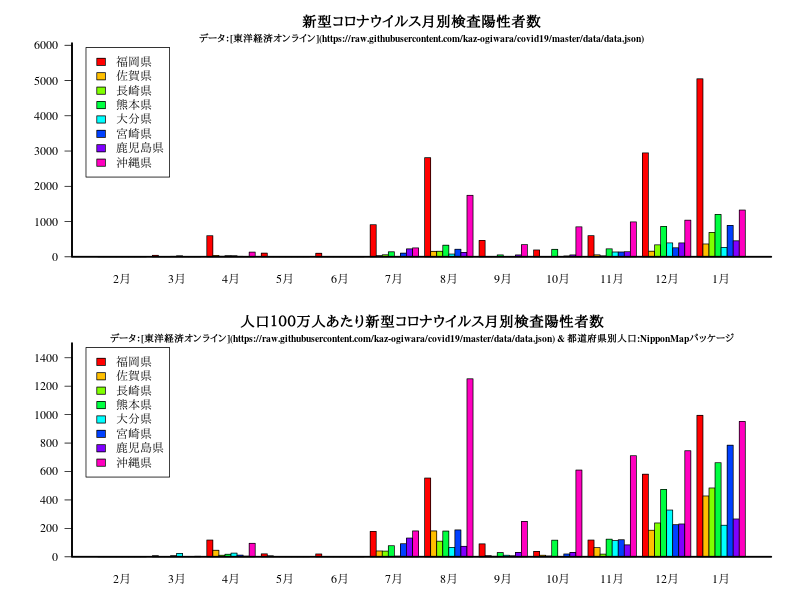
グラフ
Rコード
データ読み込み
library(jsonlite)
library(xts)
library(sf)
library(NipponMap)
#
#「東洋経済オンライン」新型コロナウイルス 国内感染の状況
# https://toyokeizai.net/sp/visual/tko/covid19/
#著作権「東洋経済オンライン」
covid19 = fromJSON("https://raw.githubusercontent.com/kaz-ogiwara/covid19/master/data/data.json")
#
names(covid19[[4]])
#[1] "carriers" "pcrtested" "discharged" "serious" "deaths"
#[6] "reproduction"
#covid19[[4]][code1,1]
#
#都道府県別人口はNipponMapパッケージのデータを使う
shp <- system.file("shapes/jpn.shp", package = "NipponMap")[1]
m <- sf::read_sf(shp)
#データの順序が一致しているか確認
covid19[[5]]$en==m$name
all(covid19[[5]]$en==m$name)
#[1] TRUE <- データの順序は一致している
#
unique(m$region)
#[1] "Hokkaido" "Tohoku" "Kanto" "Chubu"
#[5] "Kinki" "Chugoku" "Shikoku" "Kyushu / Okinawa"検査陽性者数
pngファイルで保存
name<- "carriersR"
region<- c("Hokkaido|Tohoku","Kanto","Chubu","Kinki","Chugoku|Shikoku","Kyushu / Okinawa")
#
for (r in 1:6){
code<- as.numeric(m[grep(region[r],m$region),]$SP_ID)
data<- covid19[[4]]$carriers[code[1],]
from<- as.Date(paste0(data$from[[1]][1],"-",data$from[[1]][2],"-",data$from[[1]][3]))
data.xts<- xts(x=data$values[[1]],seq(as.Date(from),length=nrow(data$values[[1]]),by="days"))
#
for (i in code[-1]){
data<- covid19[[4]]$carriers[i,]
from<- as.Date(paste0(data$from[[1]][1],"-",data$from[[1]][2],"-",data$from[[1]][3]))
tmp.xts<- xts(x=data$values[[1]],seq(as.Date(from),length=nrow(data$values[[1]]),by="days"))
data.xts<- merge(data.xts,tmp.xts)
}
# NA<- 0
coredata(data.xts)[is.na(data.xts)]<- 0
colnames(data.xts)<- covid19[[5]]$ja[code]
#
monthsum<- NULL
for (i in 1:ncol(data.xts)){
#各月ごとの検査陽性者数の合計
m.xts<- apply.monthly(data.xts[,i],sum)
monthsum<- cbind(monthsum,m.xts)
}
#
monthsum<- data.frame(monthsum)
rownames(monthsum)<- paste0(sub("^0","",substring(rownames(monthsum),6,7)),"月")
#
#if (rownames(monthsum)[nrow(monthsum)]!="11"){
# monthsum= rbind(monthsum,0)
#}
# 最初の月の検査陽性者数がすべての県で0なら削除
if ( all(monthsum[1,]==0) ) {monthsum<- monthsum[-1,]}
#plot
png(paste0(name,r,".png"),width=800,height=600)
par(mar=c(3,5,3,2),family="serif",mfrow=c(2,1))
b<- barplot(t(monthsum),beside=T,names=rownames(monthsum),las=1,col=rainbow(ncol(monthsum)),ylim=c(0,max(monthsum)*1.2),
legend=T,args.legend=list(x="topleft",inset=0.02))
box(bty="l",lwd=2.5)
#for (i in 1:ncol(monthsum)){
# text(x=b[i,],y=monthsum[,i],labels=monthsum[,i],pos=3)
#}
title("新型コロナウイルス月別検査陽性者数")
title("\n\n\nデータ:[東洋経済オンライン](https://raw.githubusercontent.com/kaz-ogiwara/covid19/master/data/data.json)",cex.main=0.8)
#
CperPop<- monthsum
for (i in 1:ncol(CperPop)){
CperPop[,i]<- round(CperPop[,i]/m$population[code[i]]*10^6,0)
}
#
#plot
par(mar=c(3,5,3,2),family="serif")
b<- barplot(t(CperPop),beside=T,names=rownames(CperPop),las=1,col=rainbow(ncol(CperPop)),ylim=c(0,max(CperPop)*1.2),
legend=T,args.legend=list(x="topleft",inset=0.02))
box(bty="l",lwd=2.5)
#for (i in 1:ncol(CperPop)){
# text(x=b[i,],y=CperPop[,i],labels=CperPop[,i],pos=3)
#}
title("人口100万人あたり新型コロナウイルス月別検査陽性者数")
title("\n\n\nデータ:[東洋経済オンライン](https://raw.githubusercontent.com/kaz-ogiwara/covid19/master/data/data.json) & 都道府県別人口:NipponMapパッケージ",cex.main=0.8)
#
dev.off()
}
par(mfrow=c(1,1))死亡者数
pngファイルで保存
name<- "deathsR"
region<- c("Hokkaido|Tohoku","Kanto","Chubu","Kinki","Chugoku|Shikoku","Kyushu / Okinawa")
#
for (r in 1:6){
code<- as.numeric(m[grep(region[r],m$region),]$SP_ID)
data<- covid19[[4]]$deaths[code[1],]
from<- as.Date(paste0(data$from[[1]][1],"-",data$from[[1]][2],"-",data$from[[1]][3]))
data.xts<- xts(x=data$values[[1]],seq(as.Date(from),length=nrow(data$values[[1]]),by="days"))
#
for (i in code[-1]){
data<- covid19[[4]]$deaths[i,]
from<- as.Date(paste0(data$from[[1]][1],"-",data$from[[1]][2],"-",data$from[[1]][3]))
tmp.xts<- xts(x=data$values[[1]],seq(as.Date(from),length=nrow(data$values[[1]]),by="days"))
data.xts<- merge(data.xts,tmp.xts)
}
# NA<- 0
coredata(data.xts)[is.na(data.xts)]<- 0
colnames(data.xts)<- covid19[[5]]$ja[code]
#
monthsum<- NULL
for (i in 1:ncol(data.xts)){
#各月ごとの死亡者の合計
m.xts<- apply.monthly(data.xts[,i],sum)
monthsum<- cbind(monthsum,m.xts)
}
#
monthsum<- data.frame(monthsum)
rownames(monthsum)<- paste0(sub("^0","",substring(rownames(monthsum),6,7)),"月")
#
#if (rownames(monthsum)[nrow(monthsum)]!="11"){
# monthsum= rbind(monthsum,0)
#}
# 最初の月の死亡者がすべての県で0なら削除
if ( all(monthsum[1,]==0) ) {monthsum<- monthsum[-1,]}
#plot
png(paste0(name,r,".png"),width=800,height=600)
par(mar=c(3,5,3,2),family="serif",mfrow=c(2,1))
b<- barplot(t(monthsum),beside=T,names=rownames(monthsum),las=1,col=rainbow(ncol(monthsum)),ylim=c(0,max(monthsum)*1.2),
legend=T,args.legend=list(x="topleft",inset=0.02))
box(bty="l",lwd=2.5)
#for (i in 1:ncol(monthsum)){
# text(x=b[i,],y=monthsum[,i],labels=monthsum[,i],pos=3)
#}
title("新型コロナウイルス月別死亡者")
title("\n\n\nデータ:[東洋経済オンライン](https://raw.githubusercontent.com/kaz-ogiwara/covid19/master/data/data.json)",cex.main=0.8)
#
CperPop<- monthsum
for (i in 1:ncol(CperPop)){
CperPop[,i]<- round(CperPop[,i]/m$population[code[i]]*10^6,0)
}
#
#plot
par(mar=c(3,5,3,2),family="serif")
b<- barplot(t(CperPop),beside=T,names=rownames(CperPop),las=1,col=rainbow(ncol(CperPop)),ylim=c(0,max(CperPop)*1.2),
legend=T,args.legend=list(x="topleft",inset=0.02))
box(bty="l",lwd=2.5)
#for (i in 1:ncol(CperPop)){
# text(x=b[i,],y=CperPop[,i],labels=CperPop[,i],pos=3)
#}
title("人口100万人あたり新型コロナウイルス月別死亡者")
title("\n\n\nデータ:[東洋経済オンライン](https://raw.githubusercontent.com/kaz-ogiwara/covid19/master/data/data.json) & 都道府県別人口:NipponMapパッケージ",cex.main=0.8)
#
dev.off()
}
par(mfrow=c(1,1))重症者数の推移
pngファイルで保存
name<- "seriousR"
region<- c("Hokkaido|Tohoku","Kanto","Chubu","Kinki","Chugoku|Shikoku","Kyushu / Okinawa")
#
for (r in 1:6){
code<- as.numeric(m[grep(region[r],m$region),]$SP_ID)
data<- covid19[[4]]$serious[code[1],]
from<- as.Date(paste0(data$from[[1]][1],"-",data$from[[1]][2],"-",data$from[[1]][3]))
data.xts<- xts(x=cumsum(data$values[[1]]),seq(as.Date(from),length=nrow(data$values[[1]]),by="days"))
#
for (i in code[-1]){
data<- covid19[[4]]$serious[i,]
from<- as.Date(paste0(data$from[[1]][1],"-",data$from[[1]][2],"-",data$from[[1]][3]))
tmp.xts<- xts(x=cumsum(data$values[[1]]),seq(as.Date(from),length=nrow(data$values[[1]]),by="days"))
data.xts<- merge(data.xts,tmp.xts)
}
# NA<- 0
coredata(data.xts)[is.na(data.xts)]<- 0
colnames(data.xts)<- covid19[[5]]$ja[code]
#
#plot
png(paste0(name,r,".png"),width=800,height=600)
par(mar=c(3,5,3,2),family="serif",mfrow=c(1,1))
matplot(coredata(data.xts),type="l",lwd=2,lty=1,las=1,col=rainbow(ncol(data.xts),alpha=0.8),
xlab="",ylab="",ylim=c(0,max(data.xts)*1.2),xaxt="n",bty="n")
box(bty="l",lwd=2.5)
labels<- sub("-","/",sub("-0","-",sub("^0","",sub("2020-","",index(data.xts)))))
labelpos<- paste0(rep(1:12,each=3),"/",1)
for (i in labelpos){
at<- match(i,labels)
if (!is.na(at)){ axis(1,at=at,labels = i,tck= -0.02)}
}
labelpos<- paste0(rep(1:12,each=3),"/",c(10,20))
for (i in labelpos){
at<- match(i,labels)
if (!is.na(at)){ axis(1,at=at,labels =NA,tck= -0.01)}
}
#都道府県名が3文字の場合全角スペースを加える
for (i in 1:ncol(data.xts)){
if (nchar(colnames(data.xts))[i]==3){
colnames(data.xts)[i]<- paste0(colnames(data.xts)[i]," ")
}
}
legend(x="topleft",inset=0.02,legend=paste0(colnames(data.xts),sprintf("%5d",tail(data.xts,1))),lwd=2,lty=1,
col=rainbow(ncol(data.xts),alpha=0.8),title=paste(index(tail(data.xts,1)),"現在"),cex=1.5,bg=rgb(0,0,0,0))
title("新型コロナウイルス重症者数の推移")
title("\n\n\nデータ:[東洋経済オンライン](https://raw.githubusercontent.com/kaz-ogiwara/covid19/master/data/data.json)",cex.main=0.8)
#
dev.off()
}表
pngファイルで保存
# webshot::install_phantomjs()
library(flextable)
library(tibble)
library(webshot)
#
zyoukyo<- data.frame(人口=formatC(m$population,format="d",big.mark=","),
PCR検査数=formatC(sapply(covid19[[4]]$pcrtested$values,sum),format="d",big.mark=","),
検査陽性者数=formatC(sapply(covid19[[4]]$carriers$values,sum),format="d",big.mark=","),
退院者数=formatC(sapply(covid19[[4]]$discharged$values,sum),format="d",big.mark=","),
死亡者数=formatC(sapply(covid19[[4]]$deaths$values,sum),format="d",big.mark=","),
重症者数=formatC(sapply(covid19[[4]]$serious$values,sum),format="d",big.mark=","))
rownames(zyoukyo)<- covid19[[5]]$ja
#knitr::kable(zyoukyo)
#
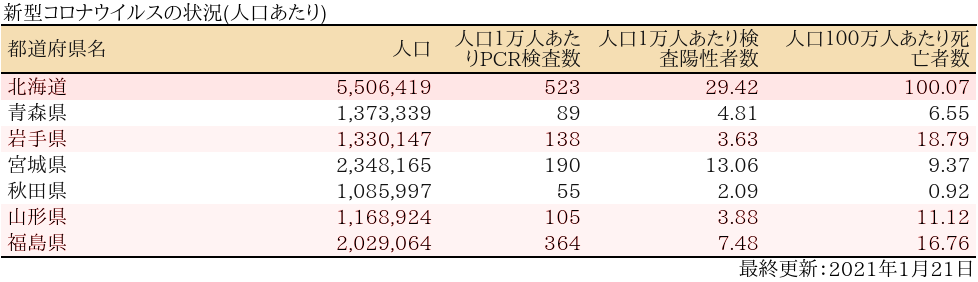
zyoukyo2<- data.frame(人口=formatC(m$population,format="d",big.mark=","),
人口1万人あたりPCR検査数=formatC(round(sapply(covid19[[4]]$pcrtested$values,sum)/m$population*10^4,0),format="d",big.mark=","),
人口1万人あたり検査陽性者数=round(sapply(covid19[[4]]$carriers$values,sum)/m$population*10^4,2),
人口100万人あたり死亡者数=round(sapply(covid19[[4]]$deaths$values,sum)/m$population*10^6,2))
rownames(zyoukyo2)<- covid19[[5]]$ja
#knitr::kable(zyoukyo2)
#
name<- "Covidtable"
region<- c("Hokkaido|Tohoku","Kanto","Chubu","Kinki","Chugoku|Shikoku","Kyushu / Okinawa")
#
for (r in 1:6){
code<- as.numeric(m[grep(region[r],m$region),]$SP_ID)
ft <- flextable(rownames_to_column(zyoukyo[code,]))
ft <- set_header_labels(ft, rowname = "都道府県名")
ft <- bg(ft, bg = "wheat", part = "header")
ft<- align(ft, i = NULL, j = -1, align = "right",part="all")
ft<- add_header_lines(ft, values = "新型コロナウイルスの状況")
ft<- add_footer_lines(ft, values =covid19[[1]]$last$ja)
ft<- align(ft, i = NULL, j = NULL, align = "right",part="footer")
# 'all', 'body', 'header', 'footer')
ft <- fontsize(ft, size = 20, part = "all")
#ft <- autofit(ft)
ft<- set_table_properties(ft, width = 1, layout = "autofit")
#ft
save_as_image(ft, path = paste0(name,r,".png"), zoom = 1, expand = 1, webshot = "webshot")
#
ft <- flextable(rownames_to_column(zyoukyo2[code,]))
ft <- set_header_labels(ft, rowname = "都道府県名")
ft <- bg(ft, bg = "wheat", part = "header")
ft <- bg(ft, i= ~人口100万人あたり死亡者数 > 10, bg = rgb(1,0,0,alpha=0.05), part = "body")
ft <- bg(ft, i= ~人口100万人あたり死亡者数 > 30, bg = rgb(1,0,0,alpha=0.1), part = "body")
ft <- color(ft, i= ~人口100万人あたり死亡者数 > 10, color = "black", part = "body")
ft<- align(ft, i = NULL, j = -1, align = "right",part="all")
ft<- add_header_lines(ft, values = "新型コロナウイルスの状況(人口あたり)")
ft<- add_footer_lines(ft, values =covid19[[1]]$last$ja)
ft<- align(ft, i = NULL, j = NULL, align = "right",part="footer")
ft <- fontsize(ft, size = 20, part = "all")
#ft <- autofit(ft)
ft<- set_table_properties(ft, width = 1, layout = "autofit")
#ft
save_as_image(ft, path = paste0(name,"2",r,".png"), zoom = 1, expand = 1, webshot = "webshot")
}